Contribution of IMGGE researchers to the fight against COVID-19
As of April 22, 11 IMGGE researchers have been engaged in COVID-19 virus testing within the newly established Huo-Yan Laboratory for Molecular Detection of Infectious Agents.
Researchers from IMGGE, along with colleagues from other scientific institutes (IBISS, IMI, INEP, INN Vinca) and faculties (Faculty of Biology, School of Medicine, Faculty of Veterinary Medicine) work in teams of ten researchers in three shifts. The laboratory with the capacity of 2000 samples per day will operate continuously during the pandemic..

A group of researchers from IMGGE have published the paper in the Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases, entitled “Functional prediction and comparative population analysis of variants in genes for proteases and innate immunity related to SARS-CoV-2 infection”.
Abstract
Functional prediction and comparative population analysis of variants in genes for proteases and innate immunity related to SARS-CoV-2 infection
New coronavirus SARS-CoV-2 is capable to infect humans and cause a novel disease COVID-19. Aiming to understand a host genetic component of COVID-19, we focused on variants in genes encoding proteases and genes involved in innate immunity that could be important for susceptibility and resistance to SARS-CoV-2 infection.
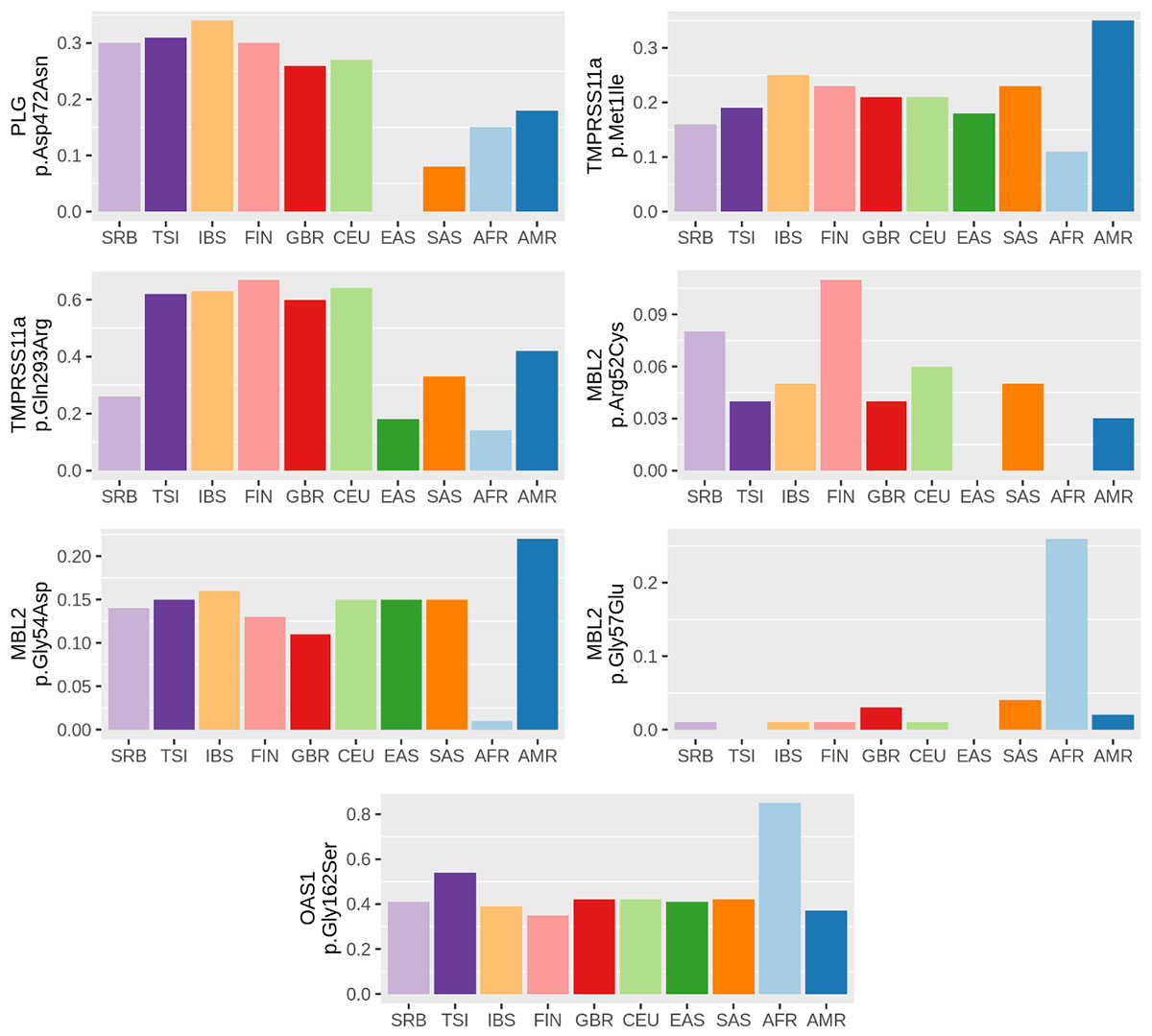
Analysis of sequence data of coding regions of FURIN, PLG, PRSS1, TMPRSS11a, MBL2 and OAS1 genes in 143 unrelated individuals from Serbian population identified 22 variants with potential functional effect. In silico analyses (PolyPhen-2, SIFT, MutPred2 and Swiss-Pdb Viewer) predicted that 10 variants could impact the structure and/or function of proteins. These protein-altering variants (p.Gly146Ser in FURIN; p.Arg261His and p.Ala494Val in PLG; p.Asn54Lys in PRSS1; p.Arg52Cys, p.Gly54Asp and p.Gly57Glu in MBL2; p.Arg47Gln, p.Ile99Val and p.Arg130His in OAS1) may have predictive value for inter-individual differences in the response to the SARS-CoV-2 infection.
Next, we performed comparative population analysis for the same variants using extracted data from the 1000 genomes project. Population genetic variability was assessed using delta MAF and Fst statistics. Our study pointed to 7 variants in PLG, TMPRSS11a, MBL2 and OAS1 genes with noticeable divergence in allelic frequencies between populations worldwide. Three of them, all in MBL2 gene, were predicted to be damaging, making them the most promising population-specific markers related to SARS-CoV-2 infection.
Comparing allelic frequencies between Serbian and other populations, we found that the highest level of genetic divergence related to selected loci was observed with African, followed by East Asian, Central and South American and South Asian populations. When compared with European populations, the highest divergence was observed with Italian population.
In conclusion, we identified 4 variants in genes encoding proteases (FURIN, PLG and PRSS1) and 6 in genes involved in the innate immunity (MBL2 and OAS1) that might be relevant for the host response to SARS-CoV-2 infection.
Pharmacogenomics landscape of COVID-19 therapy response in Serbian population and comparison with worldwide populations
Abstract
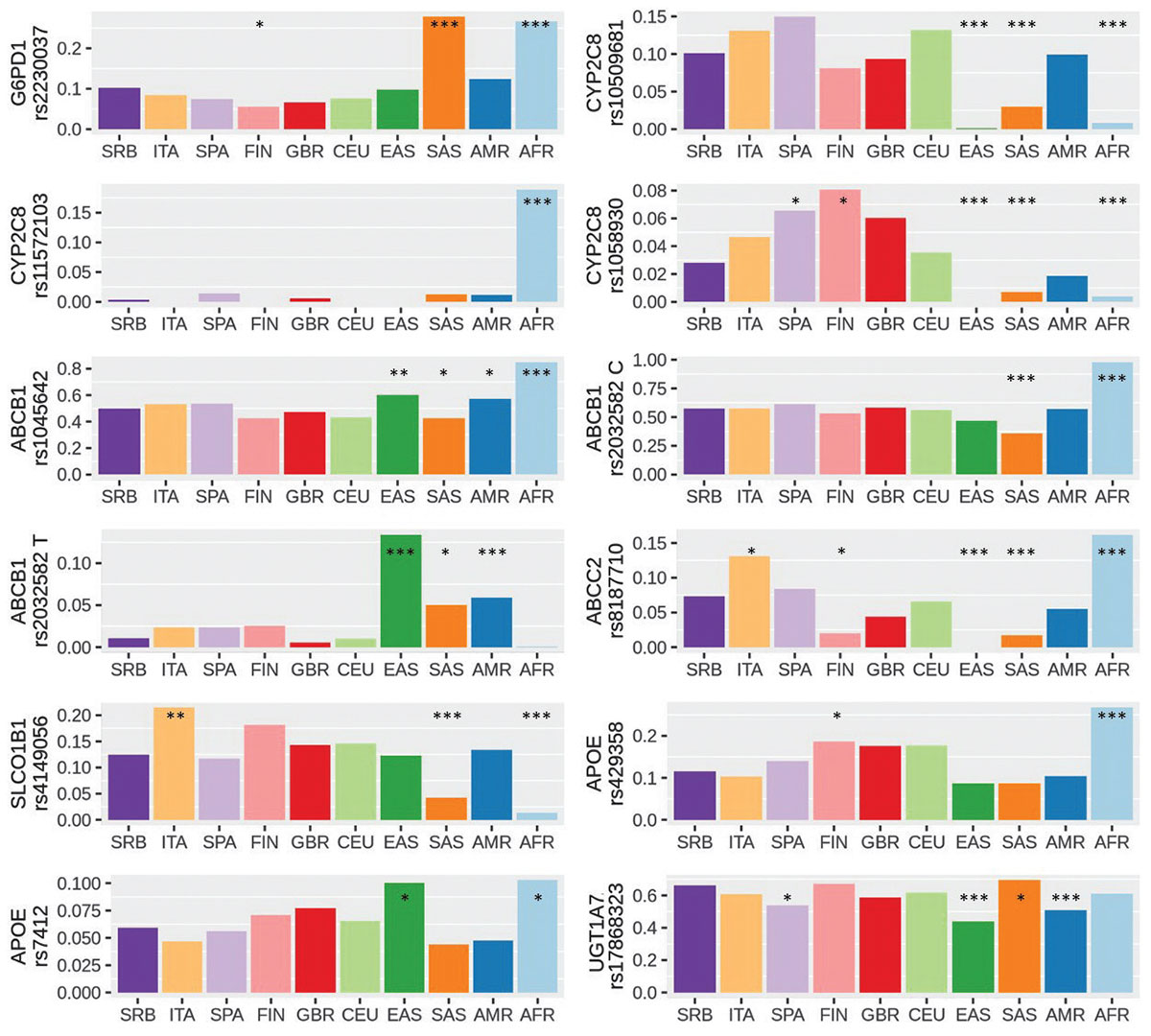
Background: Since there are no certified therapeutics to treat COVID-19 patients, drug repurposing became important. With lack of time to test individual pharmacogenomics markers, population pharmacogenomics could be helpful in predicting a higher risk of developing adverse reactions and treatment failure in COVID-19 patients. Aim of our study was to identify pharmacogenes and pharmacogenomics markers associated with drugs recommended for COVID-19 treatment, chloroquine/hydroxychloroquine, azithromycin, lopinavir and ritonavir, in population of Serbia and other world populations.
Methods: Genotype information of 143 individuals of Serbian origin was extracted from database previously obtained using TruSight One Gene Panel (Illumina). Genotype data of individuals from different world populations were extracted from the 1000 Genome Project. Fisher’s exact test was used for comparison of allele frequencies.
Results: We have identified 11 potential pharmacogenomics markers in 7 pharmacogenes relevant for COVID-19 treatment. Based on high alterative allele frequencies in population and the functional effect of the variants, ABCB1 rs1045642 and rs2032582 could be relevant for reduced clearance of azithromycin, lopinavir and ritonavir drugs and UGT1A7 rs17868323 for hyperbilirubinemia in ritonavir treated COVID-19 patients in Serbian population. SLCO1B1 rs4149056 is a potential marker of lopinavir response, especially in Italian population. Our results confirmed that pharmacogenomics profile of African population is different from the rest of the world.
Conclusions: Considering population specific pharmacogenomics landscape, preemptive testing for pharmacogenes relevant for drugs used in COVID-19 treatment could contribute to better understanding of the inconsistency in therapy response and could be applied to improve the outcome of the COVID-19 patients.
Researchers from Laboratory for Molecular Biomedicine, IMGGE participate in The COVID-19 Host Genetics Initiative (www.covid19hg.org), an international project that brings together 134 studies from 50 countries and boost them to generate, share, and analyze data to learn the genetic determinants of COVID-19 susceptibility, severity, and outcomes. Such discoveries could help to generate hypotheses for drug repurposing, identify individuals at unusually high or low risk, and contribute to global knowledge of the biology of SARS-CoV-2 infection and disease. The IMGGE team will contribute to COVID-19 HGI project with the following study:
COVID-19 response improvement through host genomic and epigenomic profiling and bioinformatics
Serbian biobank of samples accompanied with clinical data of COVID-19 patients has been established (adult, pediatric). Aim are to elucidate the role of host factors through the analysis of association of patients’ genomic and epigenomic profile with severity of COVID-19 and to investigate the age-related COVID-19 outcome. WGS of the samples, association studies and bioinformatic analysis are planned to reveal population-specific and inter-individual genomic variants related to SARS-CoV-2 infection. Investigation of the age-related COVID-19 outcome by comparison of pediatric and adult patients’ epigenomic landscape is planned. Prediction models using machine learning algorithms will be designed.
Reduced Expression of Autophagy Markers and Expansion of Myeloid-Derived Suppressor Cells Correlate With Poor T Cell Response in Severe COVID-19 Patients.
Front Immunology
Tomić S, Đokić J, Stevanović D, Ilić N, Gruden-Movsesijan A, Dinić M, Radojević D, Bekić M, Mitrović N, Tomašević R, Mikić D, Stojanović D, Čolić M.
2021 Feb 22;12:614599. doi: 10.3389/fimmu.2021.614599. PMID: 33692788; PMCID: PMC7937809.
https://pubmed.ncbi.nlm.nih.gov/33692788/#affiliation-1
Widespread coronavirus disease (COVID)-19 is causing pneumonia, respiratory and multiorgan failure in susceptible individuals. Dysregulated immune response marks severe COVID-19, but the immunological mechanisms driving COVID-19 pathogenesis are still largely unknown, which is hampering the development of efficient treatments. In the collaboration with the researchers from the Institute for the Application of Nuclear Energy (INEP), Serbian Academy of Sciences and Arts, Clinical Hospital Center Zemun, and Military Medical Academy ~140 parameters of cellular and humoral immune response in peripheral blood of 41 COVID-19 patients and 16 age/gender-matched healthy donors were analyzed, followed by integrated correlation analyses with ~30 common clinical and laboratory parameters. We found that lymphocytopenia in severe COVID-19 patients strongly affects T, NK, and NKT cells, but not B cells and antibody production. Unlike increased activation of lymphocytes in mild COVID-19 patients, T cells in severe patients showed impaired activation and anti-viral functions, which correlated with significantly down-regulated antigen-presentation potential of monocytes, dendritic cells, and B cells. The latter phenomenon was followed by impaired autophagy and the expansion of myeloid-derived suppressor cells (MDSC). Overall, down-regulated autophagy, and the expansion of MDSC subsets could play critical roles in dysregulating T cell response in COVID-19, which could have large implications in diagnostics and design of novel therapeutics for this disease.

Association of Vitamin D, Zinc and Selenium Related Genetic Variants With COVID-19 Disease Severity.
Kotur N, Skakic A, Klaassen K, Gasic V, Zukic B, Skodric-Trifunovic V, Stjepanovic M, Zivkovic Z, Ostojic O, Stevanovic G, Lavadinovic L, Pavlovic S, Stankovic B.
Frontiers in Nutrition, 2021.
Doi: 10.3389/fnut.2021.689419. PMID: 34150833; PMCID: PMC8211741.
https://pubmed.ncbi.nlm.nih.gov/34150833/
Approximately 20% of COVID-19 patients develop severe disease characterized by severe pneumonia which can lead to lethal outcome. Although certain risk factors for severe disease are already identified, the role of nutrition and patients’ genetic background is not clear. To deal with this issue, researchers from Laboratory for Molecular Biomedicine, IMGGE in collaboration with clinicians from the Clinic of Pulmonology, Clinical Center of Serbia and Children’s Hospital for Lung Diseases and Tuberculosis, Medical Center “Dr Dragiša Mišović”, carried out the following study:
Association of Vitamin D, Zinc and Selenium Related Genetic Variants With COVID-19 Disease Severity
In this study, we analyzed nutrigenetic markers previously associated with deficiency of micronutrients important for defense against viral infections: vitamin D, zinc and selenium. Our results showed difference in nutrigenetic profile between mild to moderate and severe COVID-19 patients. In particular, the differences were noted in CYP2R1 gene (involved in vitamin D transformation into its active form) and DHCR7/NADSYN gene (involved in vitamin D synthesis upon skin exposure to sunlight). Currently, we are carrying out a study on an independent group of subjects with an aim to validate our results.
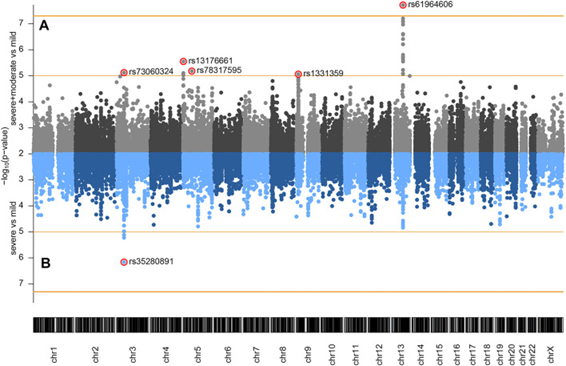
Genome-wide association study of COVID-19 outcomes reveals novel host genetic risk loci in the Serbian population
Zecevic M, Kotur N, Ristivojevic B, Gasic V, Skodric-Trifunovic V, Stjepanovic M, Stevanovic G, Lavadinovic L, Zukic B, Pavlovic S, Stankovic B.
Front Genet. 2022 Jul 14;13:911010. doi: 10.3389/fgene.2022.911010.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9329799/
We performed the first genome wide association study in Serbia related to COVID-19 as a collaborative effort of IMGGE and Clinical Centre of Serbia. DNA analysis of 128 mild, moderate and severe COVID-19 patients reveled five signals across genome as potential markers of COVID-19 severity. Our study replicated a previously reported COVID-19 risk locus at 3p21.31, identifying lead variants in SACM1L and LZTFL1 genes suggestively associated with pneumonia. Also, we have detected a significant signal associated with COVID-19 related pneumonia at locus 13q21.33, with a peak residing upstream of the gene KLHL1 associated with lung diseases. Suggestive association with COVID-19 pneumonia has also been observed at chromosomes 5p15.33, 5q11.2, and 9p23. Our results revealed novel risk loci for pneumonia and severe COVID-19 disease which could contribute to a better understanding of the COVID-19 host genetics in different populations.